-
SBDI Days
A conference to assemble and together pave the way for data-driven research in ecology. Make use of the big data wave!… [Read more].
-

Courses and training
Here you find links to training courses, online modules, and webinar recordings for education in biodiversity informatics. [Go to courses and training].
-

Ongoing research
Read about examples of active research groups and scientific networks in Sweden, which either utilize data from SBDI, or feed data into our system. [Go to ongoing research].
News
9
Apr 24
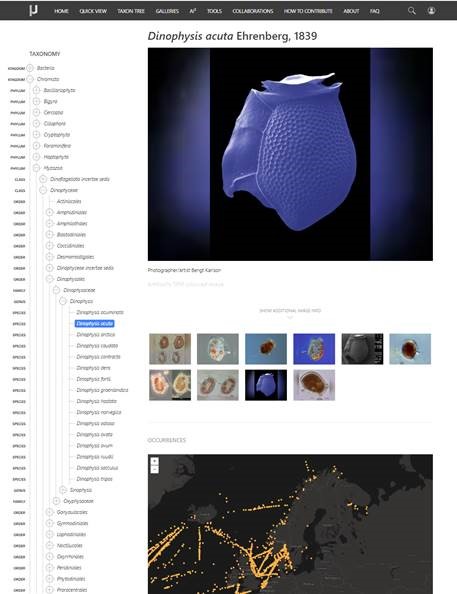
New version of Nordic Microalgae
9
Apr 24
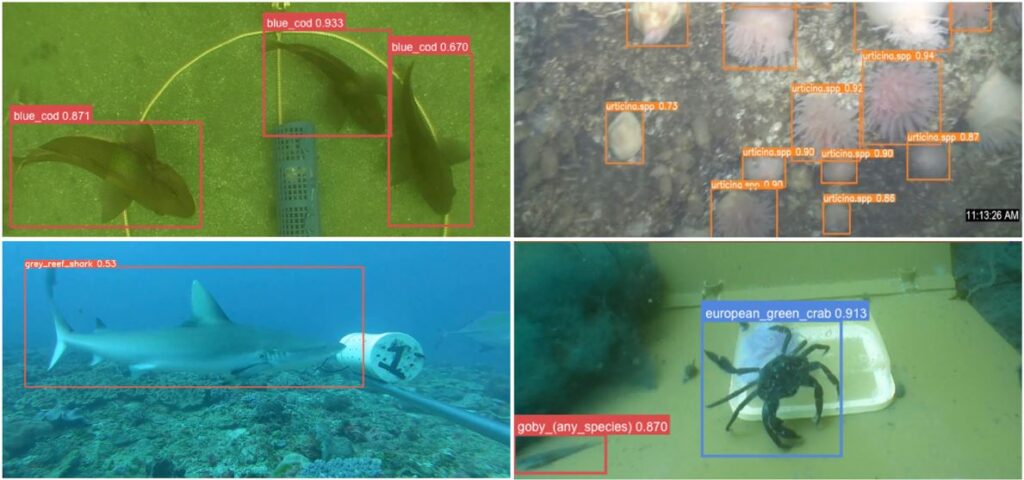
SBDI’s latest tool for image analysis
14
Mar 24
Postdoc/researcher position at Stockholm University

Events
SBDI BRINGS BIODIVERSITY DATA TOGETHER
SDBI aggregates biodiversity data from multiple sources and makes it available and usable online. It is the largest collection of freely available biodiversity data in Sweden.
MAKING THE MOST OF BIODIVERSITY DATA
Biodiversity data about plants, animals and fungi and their habitats may be used to educate and inform and in environmental decision-making, state of the environment assessments and restoration and rewilding.
.

SBDI is developed in collaboration with the Living Atlases community, and includes the Swedish node of the Global Biodiversity Information Facility (GBIF).