GS-VMAS PhD Courses
Here you find information about all our upcoming PhD courses
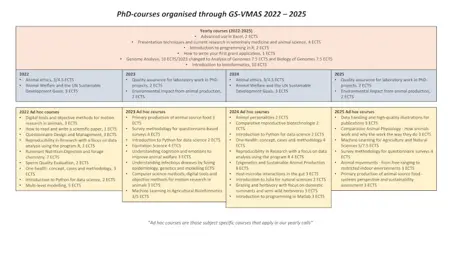
Recurring PhD courses 2026 - 2029
During 2026 - 2029 we will have a number of PhD courses that will be run on a regular basis (yearly and every second year), as shown in the picture below (to download). However, in addition to these we will also arrange a number of additional subject specific courses ("ad hoc") every year. Further information about all planned PhD courses can be found under "Upcoming PhD courses".
Upcoming PhD courses
Course code: PVG0044
Credits: 4
Course dates: Join continuously
The course has a dual format. One part is to actively participate in two workshops (4 hours per workshop) dealing with both oral and poster presentation techniques and the practical construction of a poster. Another part is to actively participate in >20 seminars given in the course seminar series. The seminar series will have weekly seminars during the spring and autumn semesters.
Course syllabus
Course code: PVG0043
Credits: 1
Course dates: 10 - 13 Feb 2026
After completing the course, you will be able to:
- Describe the different grants that are available
- Explain how to find suitable calls
- Use the information provided in the call to prepare a template for the application
- Describe what the assessors look for in the application
- Explain how to prepare a budget for a grant application
- Describe how to fill in an electronic application form
The course will consist of a series of short lectures and demonstrations, plus some exercises that the students do themselves in groups, and individually. There is a voluntary written assignment to write a grant application.
Course code: P000152
Credits: 3
Course dates: 2 - 6 March 2026 in Skara
The course integrates various disciplines, such as animal nutrition, animal welfare, animal husbandry, animal health, meat quality, economics and environmental impact on sustainable meat production under Nordic conditions. The course will deliver a holistic understanding of meat production systems in the Nordic countries with an international perspective by using interdisciplinary research in teaching.
After completing the course you should:
- be able to account for the major factors imposing constraints on the sustainability of different production systems for meat production; including theories related to nutrient partitioning and growth capacity, predicting the impact on animal production and welfare as well as on sustainability.'
- be able to discuss the major areas of concern in issues related to feed production, animal nutrition and husbandry and their impact on the environment, animal health, and meat quality.
- be able to describe results of studies about animal productivity, health, environmental and economic sustainability in different Nordic production systems.
- have gained knowledge about difference in production systems in different regions of the Nordic countries.
Tentative schedule, can be adjusted before the course start
Monday 2/3
10-12 Welcome and introduction to the course (KAS, MT) and Meat production systems (LK, MT, SE)
12-13 Lunch
13-17 Introduction to life cycle assessment (LCA) (TDP); Examples of LCA in beef production (LM) and Examples of LCA in sheep and lamb production (SS)
Evening - Pizza and short presentations of students’ projects
Tuesday 3/3
8:30-12 Meat production systems, cases and group work (SE, LK, MT)
12-13 Lunch
13-15 Visit to Götala beef and lamb research (KAS)
15-17 Introduction and start of group work, cont. Friday (ES)
Wednesday 4/3
8:30-10 Introduction to meat quality (AK, MH)
10-12 Visit to slaughter plant (ES)
12-13 Lunch
13-17 Meat quality, cases and group work (AK, MH, MT, PE)
Thursday 5/3
8:30-11 Utilization of the whole animal (MH, RR)
11-12 Industry presentation, Nortura
12-13 Lunch
Industry presentation, Danish Crown
14-16 Workshop and discussion based on industry presentations (MH, RR)
16-17 Introduction of final assignment (KAS, MT)
Evening Dinner
Friday 6/3
8:30-9:30 How to include animal welfare in sustainability assessments (EHJ)
9:30-11 Animal welfare in meat production systems (LB)
11-12 Student suggestions for a sustainable meat production in the Nordic region (ES)
12-13 Lunch
13-15 Student suggestions for a sustainable meat production in the Nordic region, cont. (ES)
15-15:30 Wrapping up and goodbye (KAS, MT)
Teachers:
AK – Anders Karlsson, Professor Emeritus, Department of Applied Animal Science and Welfare, SLU, Sweden
EHJ - Emma Hvidtfeldt Jensen, Post Doc, Department of Animal and Veterinary Science, Aarhus University, Denmark
ES – Elin Stenberg, Post Doc, Department of Applied Animal Science and Welfare, SLU, Sweden
KAS – Katarina Arvidsson Segerkvist, Associate professor, Department of Applied Animal Science and Welfare, SLU, Sweden
LB – Lotta Berg, Professor, Department of Applied Animal Science and Welfare, SLU, Sweden
LK – Liisa Keto, Senior Scientist, Production systems/animal nutrition, Natural Resources Institute Finland, Finland
LM - Lisbeth Mogensen, Senior Scientist, Department of Agroecology, Aarhus University, Denmark
MH – Marchen Hviid, Senior Scientist, Danish Technological Institute, Denmark
MT - Margrethe Therkildsen, Professor, Department of Food Science, Aarhus University, Denmark
PE – Per Ertbjerg, University Lecturer, Department of Food and Nutrition, University of Helsinki, Finland
RR – Rune Rødbotten, Scientist, Nofima, Norway
SE - Simme Eriksen, Chief Advisor Innovation Centre of Organic Agriculture, Denmark
SS – Sankalp Shrivastava, PhD student, Faculty of Food Science and Nutrition, University of Iceland
TDP - Teodora Dorca-Preda, Post Doc, Department of Agroecology, Aarhus University, Denmark
Course code: PVG0025
Credits: 2
Course dates: 16 - 27 March 2026
The course gives an introduction to R, that is a programming language for statistical computing and graphics. The course is given as an online course with recorded lectures and course meetings in zoom in addition town work with exercises.
After completing the course you should be able to:
- describe what R is and give examples on how R can be used in analysis of data
- read data into R from a file and write output to a file
- perform basic statistics and graphics in R
- use R scripts written by others and write their own shorter R scripts.
Course code: PVG0024
Credits: 2
Course dates: 7 - 21 April 2025 (three physical meetings 7 April, 14 April and 21 April (digital attendence possible))
In the course, we will go through all menus in Excel and explain how to manage data by a variety of techniques. The students will have the opportunity to increase or reduce focus on different areas. Exercises will be the main form of learning and will be conducted outside the lecture room.
On completion of the course you will be able to:
- Process research data by use of Excel "functions"
- Assure data quality
- Produce figures and tables relevant to scientific publications
- Record basic "macros"
Course code: PVG0045
Credits: 2
Course dates: 20 - 24 April 2026
After attending this course you will be able to:
- Explain basic Python syntax
- Write simple functions in Python
- Transform data in various formats using the Pandas library
- Combine data sources stored in different files using the Pandas library
- Produce insightful summaries of datasets using the Pandas library
- Produce publication quality plots using the Seaborn library
Course code: PVS0151
Credits: 2
Course dates: 18 – 21 May 2026
The course covers the following fields: the concept of sperm quality, what it is and why it is needed in different situations; how it is evaluated from basic field laboratory to advanced research setting; how it is used for different species; pitfalls in sperm quality evaluation; how it can be improved in the future. There will be a series of tutor-led lectures, workshops and practical sessions.
Course code: P000043
Credits: 7,5 credits
Course date: 31 Aug - 1 Nov 2026 (can be read parallel with Analysis of genomes (P000042))
The course intends to provide advanced knowledge of theory and methods for studies of eukaryotic genomes, including their organization and evolution. There is a focus on animal genomics, but theoretical and methodological aspects of the course are applicable on many different organisms.
The course is based on lectures, exercises, and group discussions. The contents build largely on animal genome research. Theoretical teaching is for the most part directly applicable also within for example human or plant genetics. Genome science is rapidly evolving, and the course is based on current state-of-the-art methodology and research.
Computer exercises and group discussions will cover:
- structural and comparative genomics,
- molecular evolution and phylogenetics/-genomics,
- genomic diversity and genetic variation,
- functional genomics,
- gene mapping, GWAS, and QTL analysis,
- epigenetics/-genomics,
- genome editing and transgenic animals.
The aim of the computer exercises is to give the students useful tools for basic genetic and genomic analyses. Therefore, the computer exercises use free and open-source software that the student can download and use on their own laptop. In addition to the written and the oral examination, compulsory components occur within computer exercises and group assignments.
Course code: P000042
Credits: 7,5 credits
Course date: 31 Aug - 1 nov 2026 (can be read parallel with Biology of genomes (P000043))
The course intends to provide advanced knowledge of theory and methods for analysis of eukaryotic genomes, including their organization and evolution. There is a focus on animal genomics, but theoretical and methodological aspects of the course are applicable on many different organisms.
The course is based on lectures, group discussions, exercises, and wet laboratory. The contents build largely on animal genome research. Theoretical teaching is for the most part directly applicable also within for example human or plant genetics. Genome science is rapidly evolving, and the course is based on current state-of-the-art methodology and research.
A large part of the course involves wet laboratory training including exercises and a laboratory project.
Computer exercises and group discussions will cover:
- genome sequence variation,
- genome browsing,
- primer design,
- sequence analysis,
- high throughput sequencing methodology
The aim of the computer exercises is to give the students useful tools for basic genetic and genomic analyses. Therefore, the computer exercises use free and open-source software that the student can download and use on their own laptop. In addition to the written and the oral examination, compulsory components occur within eg. exercises, group assignments and laboratory sessions.
Course code: PVS0141
Credits: 10
Course dates: 2 Nov 2026 - 17 Jan 2027
The course aims to give the student a solid foundation in the basic methods of bioinformatics. It will cover the theories, algorithms and practical applications of computer-based methods for analysis of DNA sequences and protein structures. The topics to be covered include: - biological databases - homology analysis - sequence analysis (with emphasis on the package EMBOSS and UGENE) - statistics and R introduction - web-based tools for analysis - the Unix operating system - phylogeny - comparative genomics - functional genomics - Next Generation Sequencing (NGS).
These are courses planned for 2026, but their start dates have not yet been determined. We will update the website with confirmed dates as soon as they are available.
- Simulation for Animal Breeding: Genetics, Biomarkers, and Disease transmission
- Questionnaire Design and Management
- Reproducible Research
- The infection biology, ecology and One Health perspective of vector- and reservoir-borne infectious diseases
Previous PhD Courses (2022 - 2025)
Contact
-
Graduate School for Veterinary Medicine and Animal Science (GS-VMAS)