Background
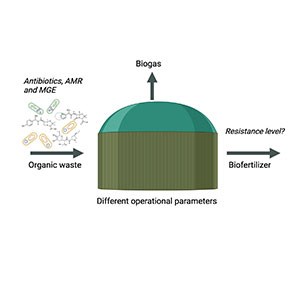
Anaerobic digestion (AD) is a well-established technology that plays a key role in developing a sustainable society. In this process, organic wastes such as animal manure, food waste and crop residues are used as substrates and converted to biogas and digestate, which represent green energy and a biofertiliser, respectively. Digestate has a high content of plant nutrients and can also contribute to soil organic carbon, which is a desirable alternative to conventional mineral fertilisers. Via organic waste materials, antibiotic resistance (AR), including both antibiotic-resistant bacteria (ARB) and antibiotic resistance genes (ARGs), can enter the biogas process. In the process, a reduction of some AR has been observed, but still, the digestate has been identified as a potential source of spreading AR.
At SLU, research is performed with the aim of assessing the levels of AR in digestate and exploring strategies for mitigating AR during the AD process. Some of the research projects are presented here.
Uncovering antibiotic resistance in biogas digestates using various substrates and mitigation strategies
Different organic waste materials can potentially contain antibiotic resistance. Today, it is unclear to what extent the resistance is reduced in the biogas process and transferred to the digestate. The resistance levels in different organic waste fractions have been evaluated through various methods, including both molecular and culture techniques. Results so far, have shown that animal manure (AM), food waste (FW) and crop residues, the main substrates for biogas production, all contain AR and that this, to some extent, is transferred to the resulting digestates. Cultivation on a selective medium to recover antibiotic resistance bacteria (ARB) shows that Bacillus and closely related genera, such as Paenibacillus and Lysinibacillus, are the dominant ARB in the digestate from both AM and FW. These bacteria displayed resistance to a broad spectrum of antibiotic classes. Additionally, ARGs and plasmids associated with Gram-negative bacteria were detected in some processes, but the results also propose a lower risk of spreading AR than substrates per se. The possibility of mitigating the risk of spreading AR is presently investigated in an ongoing postdoctoral project supported by Ekhagastiftelsen. In this project, the levels of antibiotic resistance (AR) are accessed across various stages of different biogas processes. Ten biogas plants in Sweden, utilising diverse substrates and operational methods, were chosen for evaluation. The main question to be answered is whether certain operational strategies can minimise the spreading of AR during organic farming. The study employs Resistomap's Microarray genotyping to assess AR levels and their transferability. Additionally, quantitative PCR is utilised to analyse the absolute reduction of ARGs and mobile genetic elements across the ten biogas plants.
Start and end time: 2017–2024
Funding: SLU and China Scholar Ship Council (2017-2022), Ekhaga Stiftelsen (2023-2024)
Investigation of novel resistance found in biogas digestates
Evaluating different isolates recovered from biogas digestates showed a broad resistance range during cultivation. Genomic analysis of the same isolates showed a significant mismatch between actual resistance and the presence of resistance genes. These results emphasised the need for using combined cultivation and molecular techniques for accurate assessment of resistance, but also the presence of novel resistance genes. A detailed analysis can clearly see this mismatch of one isolate, a Bacillus sp. strain FW 1, resistant to meropenem (MEM), a critical antibiotic in human medicine. Investigation into the resistance mechanism suggested the involvement of unclassified carbapenemase. The MEM-resistance in this specific bacterium was not transferable, thus representing a low risk for resistance spread when utilising digestate on arable land. However, still, the results indicate a need for further studies to fully understand the mechanism of resistance used by microbial populations in biogas processes.
Start and end time: 2017–2022)
Funding: SLU and China Scholar Ship Council
Characterisation of anaerobic antibiotic-resistant bacteria in digestates
Isolating ARB from digestate under anaerobic conditions poses a significant challenge, with limited research conducted in this area. However, anaerobic ARB potentially constitute a crucial component of the ARB community in digestate. Understanding them is essential for gaining a comprehensive insight into the ARB community, and exploring potential interactions between aerobic and anaerobic ARB is intriguing. This study seeks to devise a method for the anaerobic isolation of ARB from digestate and characterise their resistance patterns against various antibiotics.
Start and end time: 2017–2024
Funding: SLU and China Scholar Ship Council